Using McMasterPandemic
2023-01-30
1 Fast and Flexible Modelling with McMasterPandemic
McMasterPandemic is a modelling tool that was rapidly developed to provide timely insights into the Covid-19 Pandemic in Canada. We are currently refactoring this tool so that it is faster and more general. This guide describes how to use this refactored version of McMasterPandemic.
1.1 History and Motivation
1.1.1 COVID-19 Forecasts
McMasterPandemic has been used to do interesting things like predict the third wave of the COVID-19 pandemic in Ontario Canada when case numbers were going down.
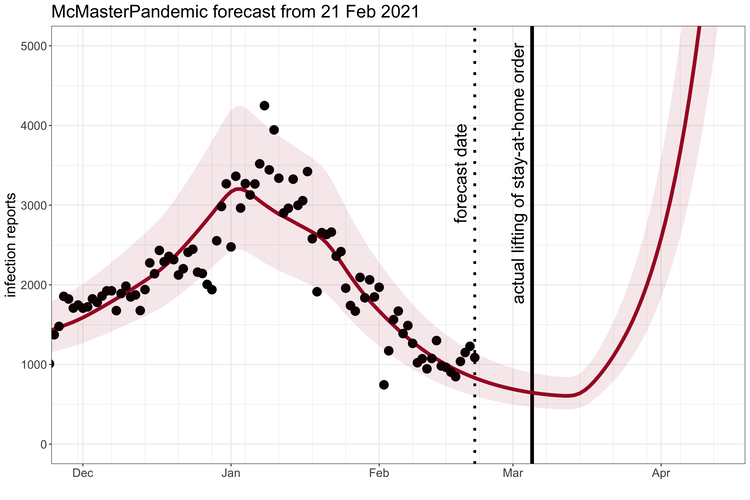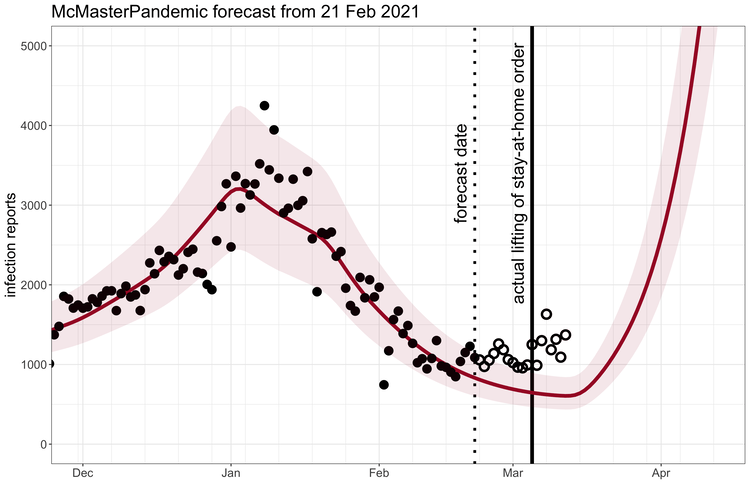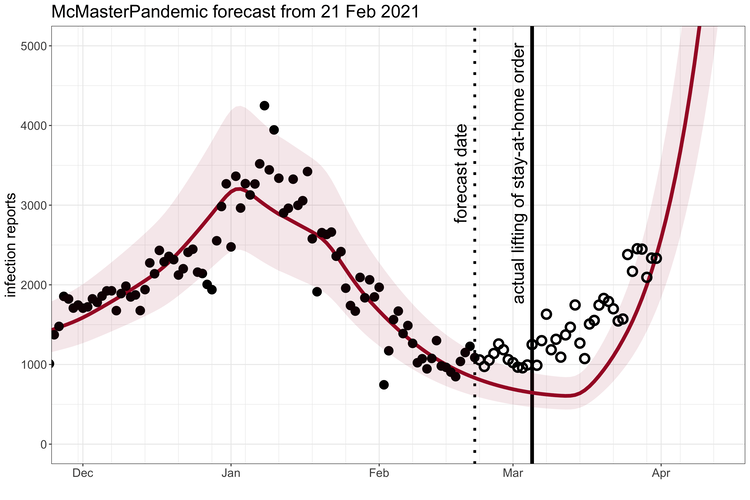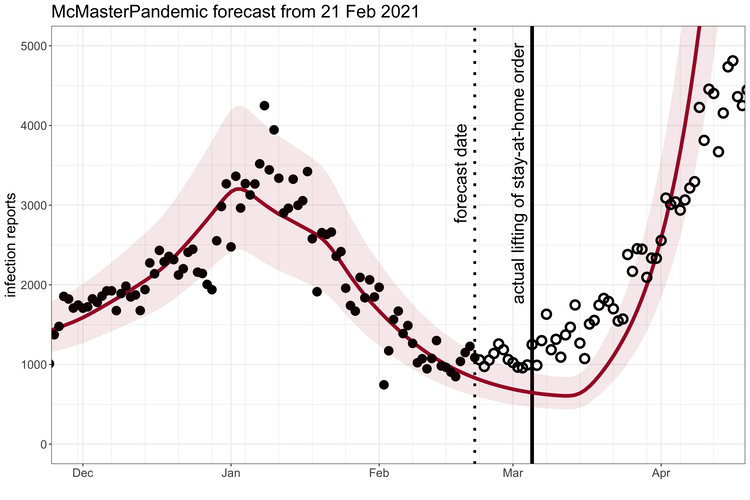
1.1.2 Calibration to data
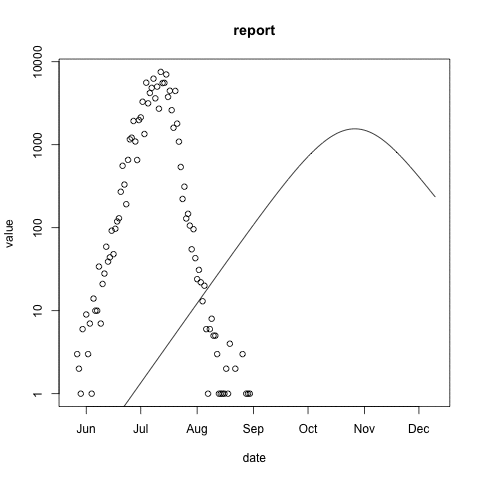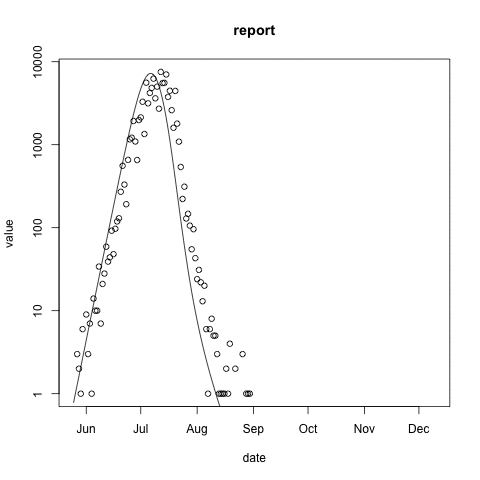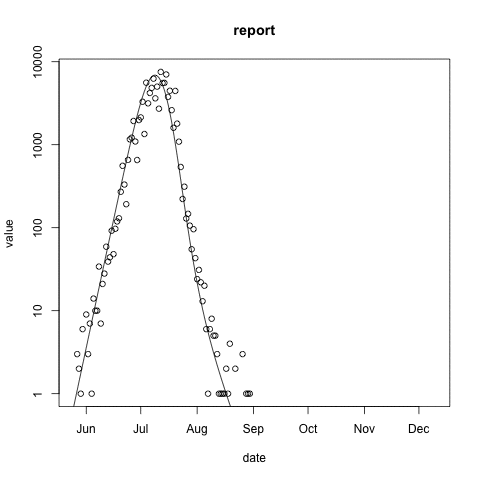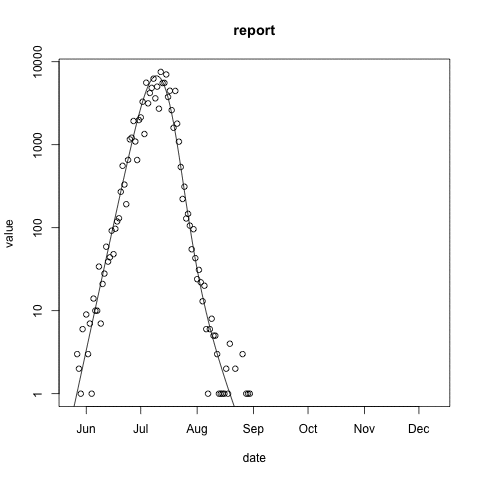
McMasterPandemic has not just been an epidemic simulation tool, but can also be used to formally calibrate to data. The following animation provides an illustrative example.
- observed case reports – dots
- simulated case reports – line
- transmission rate optimized using maximum likelihood
- negative binomial error model
1.1.3 Speed
As the COVID-19 pandemic progressed, the following scenario became more common.
- Public health modeller has to make weekly forecasts
- Model fitting/calibration takes two days
- Only three days to
- explore scenarios
- write the report
- No time to refine the model or messaging
So we refactored the McMasterPandemic engine using the TMB C++ framework, which resulted in the following unsolicited feedback from a senior research scientist at the Public Health Agency of Canada.
“It is insanely fast and definitely saves me hours everyday and allows me to explore more scenarios. I can do many more things I couldn’t do before due to computational limitations.”
1.1.4 Model Extensibility
Another common scenario also arose throughout the COVID-19 pandemic progressed.
- New variant emerges, as immunity from vaccination wanes
- Public health modeller has to create new model structure
- Requires a few weeks of software development
- Public health situation changes over this time
- Model is less relevant when complete
In response to this we built an interface allowing users to create their own compartmental model structure.
sir = (flexmodel(
params = c(beta = 0.1, gamma = 0.01, N = 100),
state = c(S = 99, I = 1, R = 0),
start_date = "2020-03-11",
end_date = "2020-12-01"
)
%>% add_rate("S", "I", ~ (I) * (beta) * (1/N))
%>% add_rate("I", "R", ~ (gamma))
)This library of models is a quick-start guide with the following examples: SIR, SI, SEIR, two-strain SIR, Erlang SEIR, SIRV, variolation SIR, SEIRD, BC’s model of the Omicron COVID variant, and Classic McMasterPandemic.
1.2 Installation
This generalized McMasterPandemic framework is still in an experimental phase. Therefore you will need to install the tmb-condense code branch that contains the experimental implementation directly from github. A convenient way to do this is to use the remotes package as follows.
1.3 Dependencies
## Warning in checkMatrixPackageVersion(): Package version inconsistency detected.
## TMB was built with Matrix version 1.5.3
## Current Matrix version is 1.5.1
## Please re-install 'TMB' from source using install.packages('TMB', type = 'source') or ask CRAN for a binary version of 'TMB' matching CRAN's 'Matrix' packageThis guide makes use of the following session.
## R version 4.2.2 (2022-10-31)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur ... 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] tmbstan_1.0.4 rstan_2.21.8 StanHeaders_2.21.0-7
## [4] bbmle_1.0.25 visNetwork_2.1.2 lubridate_1.9.1
## [7] tidyr_1.3.0 McMasterPandemic_0.2.0.0 dplyr_1.1.0
## [10] RcppEigen_0.3.3.9.3 TMB_1.9.2 ggplot2_3.4.0
##
## loaded via a namespace (and not attached):
## [1] sass_0.4.5 jsonlite_1.8.4 splines_4.2.2
## [4] foreach_1.5.2 bslib_0.4.2 RcppParallel_5.1.6
## [7] brio_1.1.3 highr_0.10 yaml_2.3.7
## [10] numDeriv_2016.8-1.1 diagram_1.6.5 pillar_1.8.1
## [13] lattice_0.20-45 glue_1.6.2 digest_0.6.31
## [16] colorspace_2.1-0 htmltools_0.5.4 Matrix_1.5-1
## [19] pkgconfig_2.0.3 bookdown_0.32 purrr_1.0.1
## [22] mvtnorm_1.1-3 scales_1.2.1 processx_3.8.0
## [25] tzdb_0.3.0 timechange_0.2.0 tibble_3.1.8
## [28] generics_0.1.3 ellipsis_0.3.2 cachem_1.0.6
## [31] withr_2.5.0 cli_3.6.0 magrittr_2.0.3
## [34] crayon_1.5.2 evaluate_0.20 ps_1.7.2
## [37] fansi_1.0.4 MASS_7.3-58.1 forcats_1.0.0
## [40] pkgbuild_1.4.0 loo_2.5.1 prettyunits_1.1.1
## [43] tools_4.2.2 hms_1.1.2 matrixStats_0.63.0
## [46] lifecycle_1.0.3 stringr_1.5.0 munsell_0.5.0
## [49] callr_3.7.3 compiler_4.2.2 jquerylib_0.1.4
## [52] rlang_1.0.6 grid_4.2.2 iterators_1.0.14
## [55] rstudioapi_0.14 htmlwidgets_1.6.1 rmarkdown_2.20
## [58] testthat_3.1.6 gtable_0.3.1 codetools_0.2-18
## [61] inline_0.3.19 R6_2.5.1 gridExtra_2.3
## [64] zoo_1.8-11 knitr_1.42 bdsmatrix_1.3-6
## [67] fastmap_1.1.0 utf8_1.2.2 fastmatrix_0.4-1245
## [70] semver_0.2.0 shape_1.4.6 readr_2.1.3
## [73] stringi_1.7.12 parallel_4.2.2 Rcpp_1.0.10
## [76] vctrs_0.5.2 tidyselect_1.2.0 xfun_0.361.4 Generalized Model at a Glance
The general model underlying McMasterPandmic’s flexible engine and interface based on a discrete time compartmental model.
\[ s_{i,t+1} = s_{i,t} + \underbrace{\sum_j M_{ji,t} s_{j,t}}_{\text{inflow}} - \underbrace{s_{i,t} \sum_j M_{ij,t} {\mathcal I}_{ij}}_{\text{outflow}} \] where,
- \(s_{i,t}\) is the state of the \(i\)th compartment at time \(t\)
- \(M_{ij,t}\) is the per-capita rate of flow from compartment \(i\) to compartment \(j\) at time \(t\)
- \({\mathcal I}_{ij}\in\{0,1\}\) indicates whether or not individuals should be removed from compartment \(i\) after flowing to compartment \(j\)
The per-capita rates, \(M_{ij,t}\), can be any expression involving only sums and products of any of the following at time \(t\):
- State of any compartment
- Model parameter, which can be time-varying – see the Model of Piece-Wise Time-Variation for details
- Complements of any of the above (i.e. \(1-x\))
- Inverses of any of the above (i.e. \(1/x\))
1.5 Vision and Direction
Our philosophy is that modellers should spend more time thinking about modelling and less time coding, troubleshooting, and waiting for computations to finish. To realize this vision we work on features that help improve at least some of the following user stories.
- Fast
- As a modeller
- I want faster forecasting and calibration
- So that I have the time to iteratively refine my models
- Rigorous
- As a scientist
- I want to use rigorous statistical techniques for fitting mechanistic models to data
- So that I can produce defensible descriptions of the processes that I am modelling
- Functionality-rich
- As a modeller
- I need to use software that provides an extensive set of modelling capabilities
- So that I can address specific and changing public health needs, which building on what I have done before
- Numerically-stable
- As a modeller
- I need software that ideally hides the details of scientific computing from me (e.g. I don’t need to worry about things like convergence) and at least provides informative and actionable messages about numerical issues
- So that I can focus on the biology and public-health implications
- Editable
- As a modeller
- I want to build complex models out of simpler – easier to understand – modular sub-models
- So that I can more easily modify the structure of my model as public health needs change
- Versionable
- As a modeller
- I want to save my model definitions in format that is both human-readable and computer-consumable
- So that I can easily see the differences between two model versions and easily review the details of models used for past forecasts
- Extensible
- As a developer
- I want to be able to quickly add a new modelling capability
- So that I can support public health modellers in as timely a manner as possible
The stories at the start of this list have been more fully realized in the existing software, and those at the bottom are more aspirational. This roadmap describes our current thoughts on what we should do next to address these five stories in as complete a manner as possible.