The vignette("quickstart") and
vignette("calibration") articles work with simulated data,
but it is more interesting to fit models to real data. The International
Infectious Disease Data Archive (IIDDA) is a useful place to look
for incidence, mortality, and population data for illustrating
macpan2. This archive contains data from this data digitization
project, which we will use here. What we find is that with real
data, even with relatively simple models and calibration problems,
issues can arise that require careful thought.
Setup
As usual we need the following packages. If you don’t have them,
please get them in the usual way (i.e. using
install.packages).
In addition, we need the iidda.api package.
if (!require(iidda.api)) {
repos = c(
"https://canmod.r-universe.dev"
, "https://cran.r-project.org"
)
install.packages("iidda.api", repos = repos)
}
api_hook = iidda.api::ops_stagingAnd we set a few options for convenience.
options(iidda_api_msgs = FALSE, macpan2_verbose = FALSE)One Scarlet Fever Outbreak in Ontario
The following code will pull data for a single scarlet fever outbreak in Ontario that ended in 1930.
scarlet_fever_ontario = api_hook$filter(
resource_type = "Compilation"
, dataset_ids = "canmod-cdi-harmonized"
, iso_3166_2 = "CA-ON" ## get ontario data
, time_scale = "wk" ## weekly incidence data only
, disease = "scarlet-fever"
# get data between 1929-08-01 and 1930-10-01
, period_end_date = "1929-08-01..1930-10-01"
)
print(scarlet_fever_ontario)
#> # A tibble: 61 × 19
#> basal_disease cases_this_period collection_year dataset_id digitization_id
#> <chr> <dbl> <dbl> <chr> <chr>
#> 1 scarlet-fever 23 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 2 scarlet-fever 27 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 3 scarlet-fever 47 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 4 scarlet-fever 24 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 5 scarlet-fever 24 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 6 scarlet-fever 45 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 7 scarlet-fever 57 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 8 scarlet-fever 46 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 9 scarlet-fever 45 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> 10 scarlet-fever 67 1929 canmod-cdi-h… cdi_sf_ca_1924…
#> # ℹ 51 more rows
#> # ℹ 14 more variables: disease <chr>, historical_disease <chr>,
#> # historical_disease_family <chr>, historical_disease_subclass <chr>,
#> # iso_3166 <chr>, iso_3166_2 <chr>, location <chr>, location_type <chr>,
#> # nesting_disease <chr>, original_dataset_id <chr>, period_end_date <date>,
#> # period_start_date <date>, scan_id <chr>, time_scale <chr>This is what the data looks like.
(scarlet_fever_ontario
|> ggplot(aes(period_end_date, cases_this_period))
+ geom_line() + geom_point()
+ ggtitle("Scarlet Fever Incidence in Ontario, Canada")
+ theme_bw()
)
SIR Model
We will begin by pulling the simple sir model from the model library.
sir = mp_tmb_library("starter_models", "sir", package = "macpan2")
print(sir)
#> ---------------------
#> Default values:
#> quantity value
#> beta 0.2
#> gamma 0.1
#> N 100.0
#> I 1.0
#> R 0.0
#> ---------------------
#>
#> ---------------------
#> Before the simulation loop (t = 0):
#> ---------------------
#> 1: S ~ N - I - R
#>
#> ---------------------
#> At every iteration of the simulation loop (t = 1 to T):
#> ---------------------
#> 1: mp_per_capita_flow(from = "S", to = "I", rate = "beta * I / N",
#> flow_name = "infection")
#> 2: mp_per_capita_flow(from = "I", to = "R", rate = "gamma", flow_name = "recovery")Modify Model for Reality
Once we take any library model off the shelf we need to modify it a
bit to be more realistic. We do this with the
mp_tmb_insert() function, which inserts new default values
and model expressions.
We first change the population size to the population of Ontario at the time.
ontario_population = api_hook$filter(
resource_type = "Compilation"
, dataset_ids = "canmod-pop-normalized"
, iso_3166_2 = "CA-ON" ## get ontario data
# get data between 1929-08-01 and 1930-10-01
, date = "1929-08-01..1930-10-01"
)
sf_sir = mp_tmb_insert(sir
, default = list(N = median(ontario_population$population))
)Second, our SIR model assumes that all of the cases are recorded in
the data, but this is not realistic. We therefore modify the model to
include under-reporting, by creating a case reports
variable that is given by the product of incidence (i.e. weekly
infection rate) and a reporting probability.
sf_sir = mp_tmb_insert(sf_sir
## insert this expression ...
, expressions = list(reports ~ infection * report_prob)
## at the end (i.e. Infinity) of the expressions evaluated
## 'during' each iteration of the simulation loop ...
, at = Inf
, phase = "during"
## add a new default value for the reporting probability
, default = list(report_prob = 1/300)
)Here we can print out the modified model to see if our changes were made successfully.
print(sf_sir)
#> ---------------------
#> Default values:
#> quantity value
#> beta 2.000000e-01
#> gamma 1.000000e-01
#> N 3.393512e+06
#> I 1.000000e+00
#> R 0.000000e+00
#> report_prob 3.333333e-03
#> ---------------------
#>
#> ---------------------
#> Before the simulation loop (t = 0):
#> ---------------------
#> 1: S ~ N - I - R
#>
#> ---------------------
#> At every iteration of the simulation loop (t = 1 to T):
#> ---------------------
#> 1: mp_per_capita_flow(from = "S", to = "I", rate = "beta * I / N",
#> flow_name = "infection")
#> 2: mp_per_capita_flow(from = "I", to = "R", rate = "gamma", flow_name = "recovery")
#> 3: reports ~ infection * report_probPreparing the Data
The first step in preparing data for macpan2 is to
simulate from the model that you are considering. Here we simulate the
"reports" variable because it corresponds to the reported
incidence (the number of new cases per time-step, which in our case is
one week, multiplied by a reporting probability).
sir_simulator = mp_simulator(sf_sir
, time_steps = 5
, outputs = "reports"
)
head(mp_trajectory(sir_simulator))
#> matrix time row col value
#> 1 reports 1 0 0 0.0006666665
#> 2 reports 2 0 0 0.0007333330
#> 3 reports 3 0 0 0.0008066662
#> 4 reports 4 0 0 0.0008873327
#> 5 reports 5 0 0 0.0009760658The next step is to get our data into a format that is compatible
with the format of these simulations. In particular, we need
matrix, time, and value columns.
We can omit the row and col columns, because
all of the ‘matrices’ in the model are 1-by-1 scalars.
observed_data = (scarlet_fever_ontario
## select the variables to be modelled -- a time-series of case reports.
|> select(period_end_date, cases_this_period)
## change the column headings so that they match the columns
## in the simulated trajectories.
|> mutate(matrix = "reports")
|> rename(value = cases_this_period)
## create a `time` column with the time-step IDs that will correspond
## to the time-steps in the simulation. this column heading also
## must match the column with the time-steps in the simulated trajectories
|> mutate(time = seq_along(period_end_date))
)
print(head(observed_data))
#> # A tibble: 6 × 4
#> period_end_date value matrix time
#> <date> <dbl> <chr> <int>
#> 1 1929-08-03 23 reports 1
#> 2 1929-08-10 27 reports 2
#> 3 1929-08-17 47 reports 3
#> 4 1929-08-24 24 reports 4
#> 5 1929-08-31 24 reports 5
#> 6 1929-09-07 45 reports 6Set up the Optimizer
Now we can create an object that can be calibrated.
sir_cal = mp_tmb_calibrator(
spec = sf_sir
, data = observed_data
## name the trajectory variable, with a name that
## is the same in both the spec and the data
, traj = "reports"
## fit the following parameters
, par = c("beta", "gamma", "I", "report_prob")
)Here we assert that we will fit beta,
gamma, (the initial value of) I, and
report_prob. Note that we can only choose to fit parameters
in the default list of the model spec. In particular,
I is in the default list because the model requires the
initial number of infectious individuals, whereas S is not
because it is derived before the simulation loop as
S ~ N - I - R.
print(sf_sir)
#> ---------------------
#> Default values:
#> quantity value
#> beta 2.000000e-01
#> gamma 1.000000e-01
#> N 3.393512e+06
#> I 1.000000e+00
#> R 0.000000e+00
#> report_prob 3.333333e-03
#> ---------------------
#>
#> ---------------------
#> Before the simulation loop (t = 0):
#> ---------------------
#> 1: S ~ N - I - R
#>
#> ---------------------
#> At every iteration of the simulation loop (t = 1 to T):
#> ---------------------
#> 1: mp_per_capita_flow(from = "S", to = "I", rate = "beta * I / N",
#> flow_name = "infection")
#> 2: mp_per_capita_flow(from = "I", to = "R", rate = "gamma", flow_name = "recovery")
#> 3: reports ~ infection * report_probRun the Optimization
sir_opt = mp_optimize(sir_cal)
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluationNot off to a good start. Those warnings are not necessarily bad, but they might get us thinking.
Examine the fit
print(sir_opt)
#> $par
#> params params params params
#> 35.6519094 35.5361402 1.9567217 0.3568965
#>
#> $objective
#> [1] 428.1962
#>
#> $convergence
#> [1] 1
#>
#> $iterations
#> [1] 145
#>
#> $evaluations
#> function gradient
#> 200 146
#>
#> $message
#> [1] "function evaluation limit reached without convergence (9)"OK, things are not great. The convergence code is
1, indicating that the model did not converge
(convergence == 0 is good). Examining the parameter
estimates, which are stored internally in the calibrator object, things
get worse.
mp_tmb_coef(sir_cal, conf.int = TRUE)
#> term mat row col default type estimate std.error
#> 1 params beta 0 0 0.200000000 fixed 35.6519094 26.8422325
#> 2 params.1 gamma 0 0 0.100000000 fixed 35.5361402 26.8423831
#> 3 params.2 I 0 0 1.000000000 fixed 1.9567217 2.9440423
#> 4 params.3 report_prob 0 0 0.003333333 fixed 0.3568965 0.2688191
#> conf.low conf.high
#> 1 -16.9578995 88.2617183
#> 2 -17.0739640 88.1462444
#> 3 -3.8134952 7.7269385
#> 4 -0.1699793 0.8837722That doesn’t look right! Those are very high beta and
gamma values, and the confidence intervals are enormous and
overlap zero.
But the model fit doesn’t look all that bad.
fitted_data = mp_trajectory_sd(sir_cal, conf.int = TRUE)
(observed_data
|> ggplot()
+ geom_point(aes(time, value))
+ geom_line(aes(time, value)
, data = fitted_data
)
+ geom_ribbon(aes(time, ymin = conf.low, ymax = conf.high)
, data = fitted_data
, alpha = 0.2
, colour = "red"
)
+ theme_bw()
+ facet_wrap(~matrix, ncol = 1, scales = "free")
)
What is going on? Can we modify this model and/or fitting procedure to get both reasonable parameter estimates and a reasonable trajectory?
Fixing the Optimization Problem
One way to address the lack of convergence of the optimizer is to
assess the degree to which the parameter estimates are biologically
reasonable. Take the recovery rate, which is estimated as
gamma ~ 26.5 per time-step. Given that a time-step is one
week in this model, this estimate implies that individuals recover from
scarlet fever in 1/26.5 weeks – much less than a day. A
quick search suggests to me that this recovery rate is not reasonable
for scarlet fever, and that a rough guess that the infectious stage
lasts one week is much more plausible than 1/26.5 weeks.
Now look at the transmission rate, beta. It is also
estimated to be pretty large, but what is more interesting is the large
standard errors for both beta and gamma. These
standard errors suggest that the estimates are not precise. However, the
estimated correlation between beta and gamma
is very close to 1, suggesting that many values for these
parameters would fit the data well as long as they are both of similar
magnitude.
mp_tmb_fixef_cov(sir_cal) |> cov2cor()
#> beta gamma I report_prob
#> beta 1.0000000 1.0000000 -0.9998612 0.9996967
#> gamma 1.0000000 1.0000000 -0.9998604 0.9996980
#> I -0.9998612 -0.9998604 1.0000000 -0.9993254
#> report_prob 0.9996967 0.9996980 -0.9993254 1.0000000This diagnosis suggests that the data are not sufficiently
informative to identify values for both beta and
gamma. Resolving such identifiability issues is often best
done by introducing prior information, such as our rough guess that
gamma is close to 1. The simplest way to
include such prior information is to move gamma out of the
pars argument to mp_tmb_calibrator and into
the default argument, as below.
sir_cal_assume_gamma = mp_tmb_calibrator(
spec = sf_sir
, data = observed_data
## name the trajectory variable, with a name that
## is the same in both the spec and the data
, traj = "reports"
## fit the following parameters
, par = c("beta", "I", "report_prob")
, default = list(gamma = 1)
)This calibration specification does not try to jointly fit both
beta and gamma, but rather fits only
beta while assuming that gamma = 1.
sir_opt_assume_gamma = mp_optimize(sir_cal_assume_gamma)
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluation
print(sir_opt_assume_gamma)
#> $par
#> params params params
#> 1.121602e+00 1.878305e+03 1.125689e-02
#>
#> $objective
#> [1] 430.0021
#>
#> $convergence
#> [1] 0
#>
#> $iterations
#> [1] 77
#>
#> $evaluations
#> function gradient
#> 121 78
#>
#> $message
#> [1] "relative convergence (4)"More warnings, but now the optimizer converges. And the standard errors in the coefficient table and fixed effect correlations seem more plausible.
mp_tmb_coef(sir_cal_assume_gamma)
#> term mat row col default type estimate std.error
#> 1 params beta 0 0 0.200000000 fixed 1.121602e+00 1.801964e-03
#> 2 params.1 I 0 0 1.000000000 fixed 1.878305e+03 5.445606e+01
#> 3 params.2 report_prob 0 0 0.003333333 fixed 1.125689e-02 1.927763e-04
mp_tmb_fixef_cov(sir_cal_assume_gamma) |> cov2cor()
#> beta I report_prob
#> beta 1.0000000 -0.7988821 -0.7608921
#> I -0.7988821 1.0000000 0.5802420
#> report_prob -0.7608921 0.5802420 1.0000000And the fit looks similar to the four-parameter model, which is consistent with our diagnosis of non-identifiability.
fitted_data = mp_trajectory_sd(sir_cal_assume_gamma, conf.int = TRUE)
(observed_data
|> ggplot()
+ geom_point(aes(time, value))
+ geom_line(aes(time, value)
, data = fitted_data
)
+ geom_ribbon(aes(time, ymin = conf.low, ymax = conf.high)
, data = fitted_data
, alpha = 0.2
, colour = "red"
)
+ theme_bw()
+ facet_wrap(~matrix, ncol = 1, scales = "free")
)
Caution: Here we use prior information to assume a reasonable value
of gamma = 1, but ignore prior uncertainty. This can cause
underestimates of uncertainty in other parameters that are fitted (see
(Elderd,
Dukic, and Dwyer 2006)). It is better to use a prior
distribution.
Learning the Functional form of Time Variation in Transmission (New!)
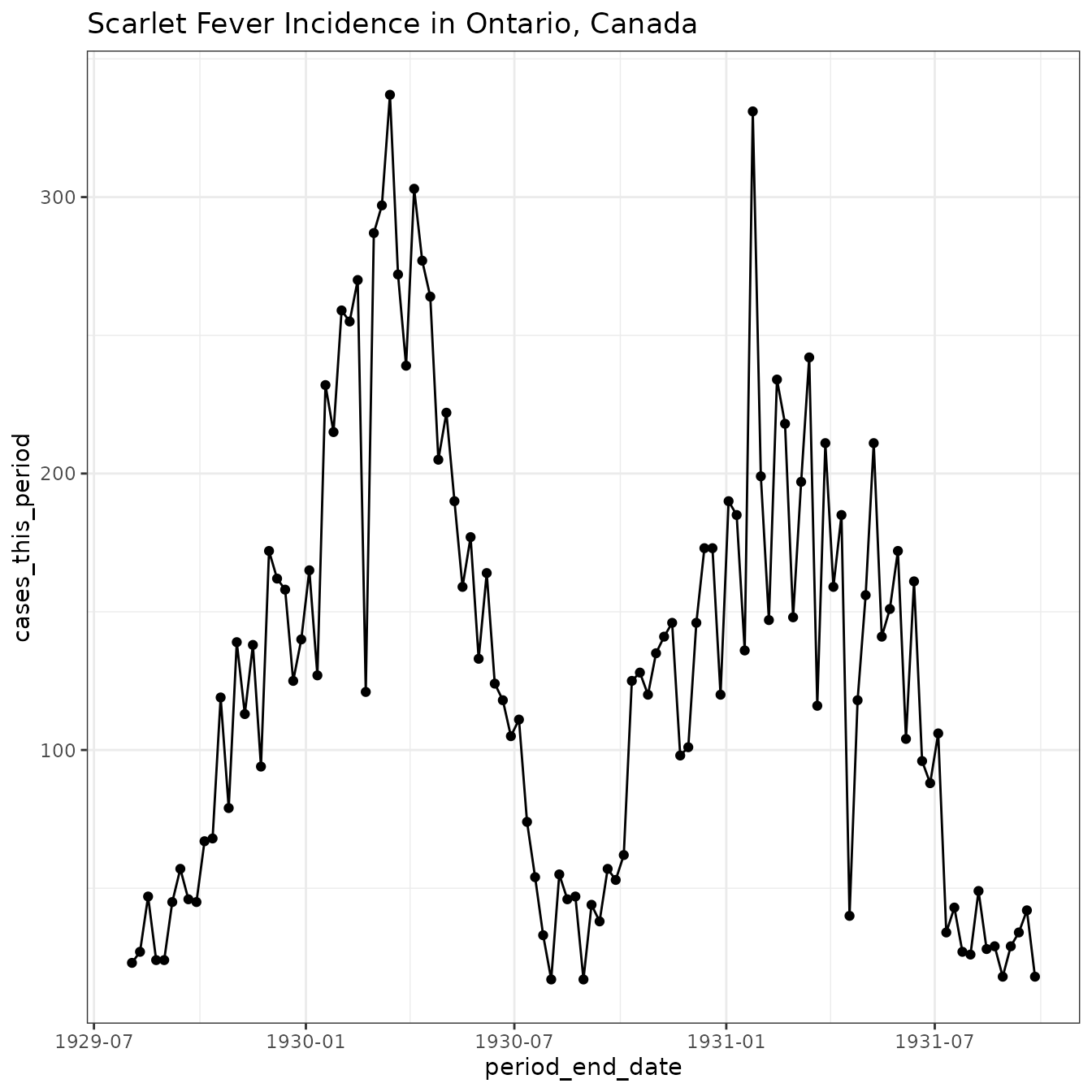
Let’s get a bit more data to see two seasons.
scarlet_fever_ontario = api_hook$filter(
resource_type = "Compilation"
, dataset_ids = "canmod-cdi-harmonized"
, iso_3166_2 = "CA-ON" ## get ontario data
, time_scale = "wk" ## weekly incidence data only
, disease = "scarlet-fever"
# get data between 1929-08-01 and 1931-10-01
, period_end_date = "1929-08-01..1931-10-01"
)
(scarlet_fever_ontario
|> ggplot(aes(period_end_date, cases_this_period))
+ geom_line() + geom_point()
+ ggtitle("Scarlet Fever Incidence in Ontario, Canada")
+ theme_bw()
)
observed_data = (scarlet_fever_ontario
## select the variables to be modelled -- a time-series of case reports.
|> select(period_end_date, cases_this_period)
## change the column headings so that they match the columns
## in the simulated trajectories.
|> mutate(matrix = "reports")
|> rename(value = cases_this_period)
## create a `time` column with the time-step IDs that will correspond
## to the time-steps in the simulation. this column heading also
## must match the column with the time-steps in the simulated trajectories
|> mutate(time = seq_along(period_end_date))
)
print(head(observed_data))
#> # A tibble: 6 × 4
#> period_end_date value matrix time
#> <date> <dbl> <chr> <int>
#> 1 1929-08-03 23 reports 1
#> 2 1929-08-10 27 reports 2
#> 3 1929-08-17 47 reports 3
#> 4 1929-08-24 24 reports 4
#> 5 1929-08-31 24 reports 5
#> 6 1929-09-07 45 reports 6Prepare to fit to the data. We make a function so that we can easily update the dimension of the radial basis.
make_rbf_calibrator = function(dimension) {
mp_tmb_calibrator(
spec = sf_sir
, data = observed_data
, traj = "reports"
## -----------------------------
## this is the key bit
, tv = mp_rbf("beta", dimension)
## -----------------------------
, par = list(
gamma = mp_unif()
, I = mp_unif()
, report_prob = mp_unif()
)
)
}And it is also convenient to make a function for plotting the results
plot_fit = function(cal_object) {
fitted_data = mp_trajectory_sd(cal_object, conf.int = TRUE)
(observed_data
|> ggplot()
+ geom_point(aes(time, value))
+ geom_line(aes(time, value)
, data = fitted_data
, colour = "red"
)
+ geom_ribbon(aes(time, ymin = conf.low, ymax = conf.high)
, data = fitted_data
, alpha = 0.2
, colour = "red"
)
+ theme_bw()
)
}Now we try fitting for a number of different dimensions.
sir_cal = make_rbf_calibrator(dimension = 1)
mp_optimize(sir_cal)
#> $par
#> params params params params params
#> 1.06263996 436.37780942 0.18345472 0.08100246 0.86580386
#>
#> $objective
#> [1] 2896.6
#>
#> $convergence
#> [1] 1
#>
#> $iterations
#> [1] 150
#>
#> $evaluations
#> function gradient
#> 168 151
#>
#> $message
#> [1] "iteration limit reached without convergence (10)"
plot_fit(sir_cal)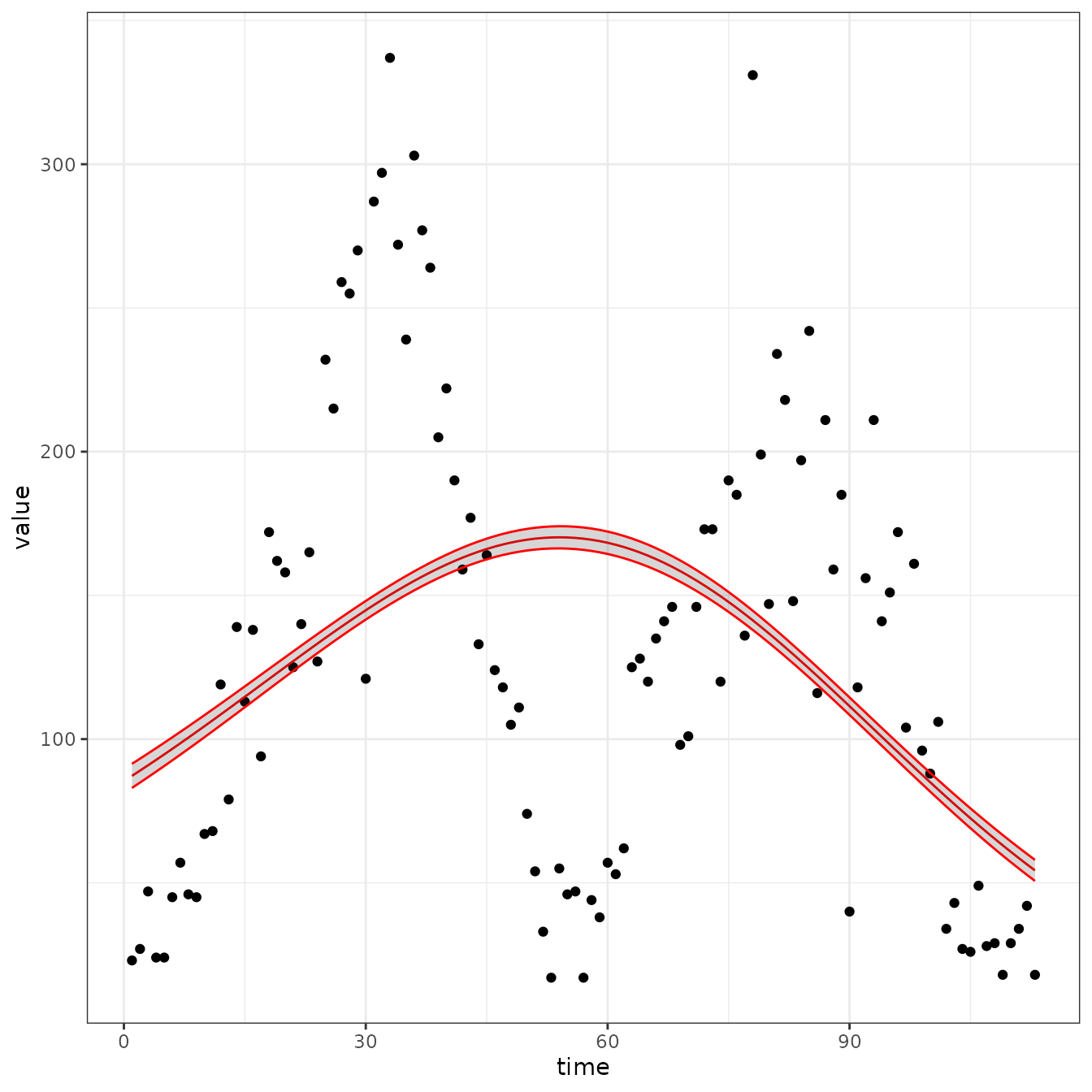
sir_cal = make_rbf_calibrator(dimension = 2)
mp_optimize(sir_cal)
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluation
#> $par
#> params params params params params params
#> 0.4658727 14.6766010 9.1431125 -0.5636248 -0.7005872 0.6673706
#>
#> $objective
#> [1] 2977.812
#>
#> $convergence
#> [1] 1
#>
#> $iterations
#> [1] 150
#>
#> $evaluations
#> function gradient
#> 198 151
#>
#> $message
#> [1] "iteration limit reached without convergence (10)"
plot_fit(sir_cal)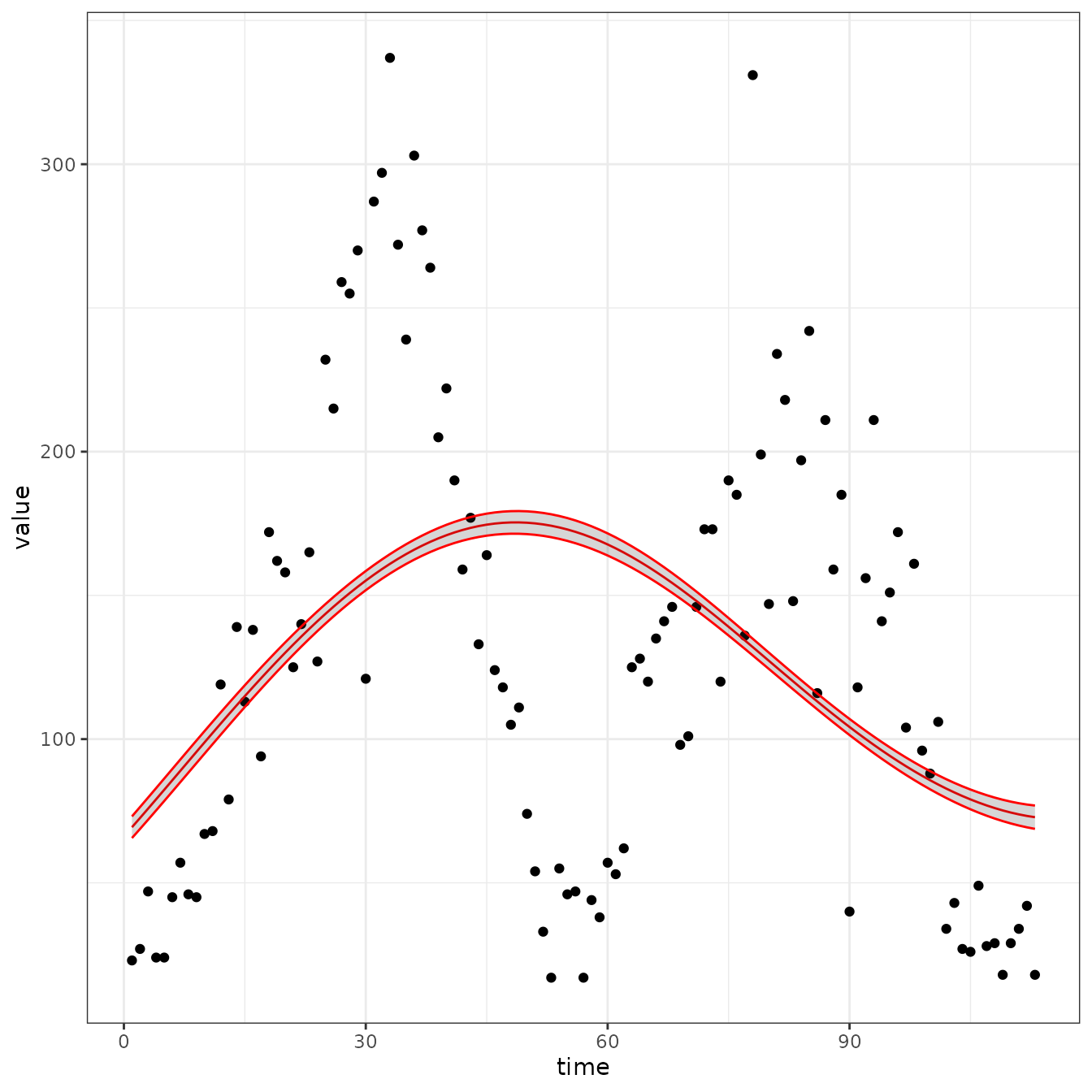
sir_cal = make_rbf_calibrator(dimension = 3)
mp_optimize(sir_cal)
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluation
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluation
#> $par
#> params params params params params
#> 7.480423e-01 7.126562e+03 5.678344e-03 -1.637587e-01 -2.970186e-02
#> params params
#> 9.244461e-01 6.246204e-01
#>
#> $objective
#> [1] 1574.227
#>
#> $convergence
#> [1] 0
#>
#> $iterations
#> [1] 67
#>
#> $evaluations
#> function gradient
#> 98 68
#>
#> $message
#> [1] "relative convergence (4)"
plot_fit(sir_cal)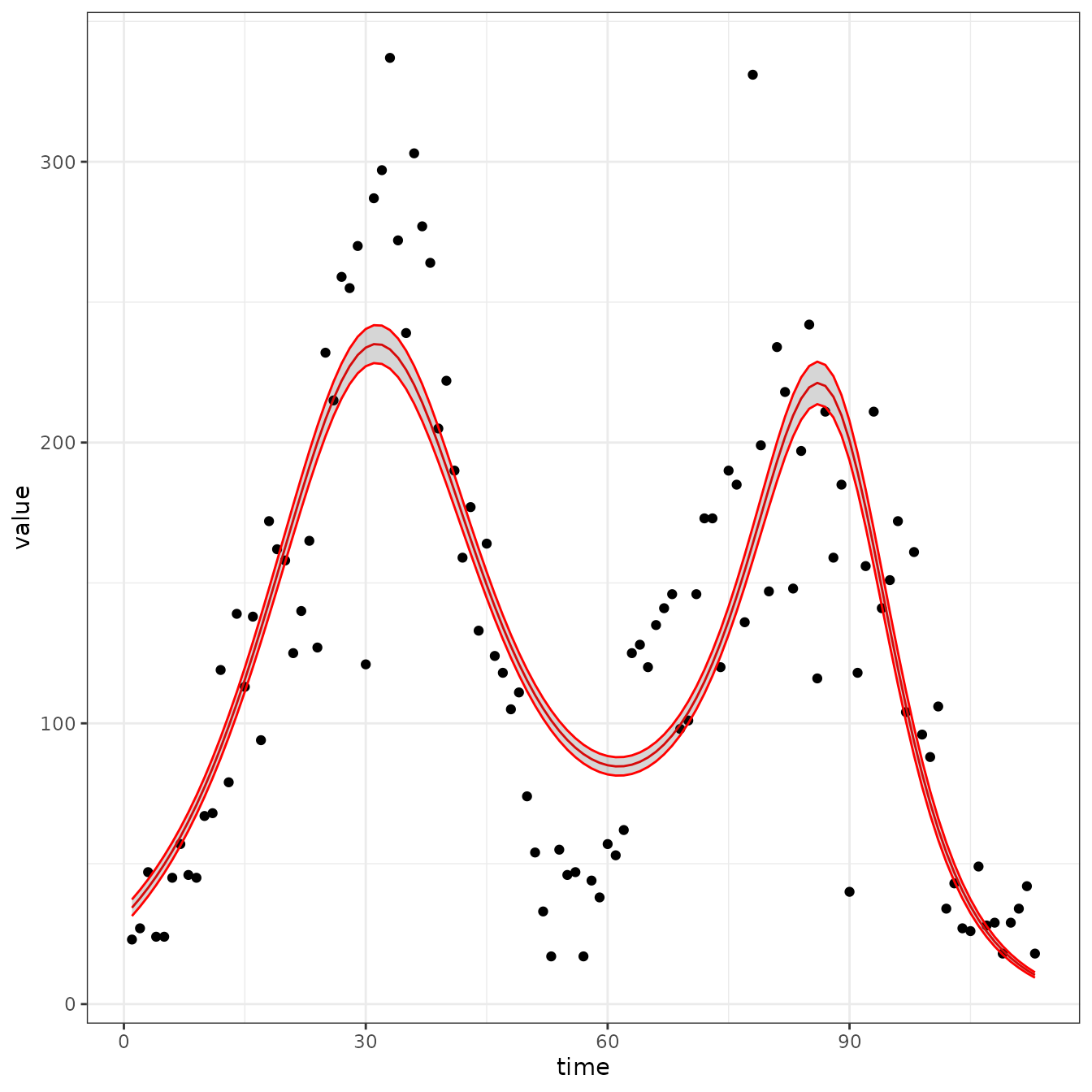
sir_cal = make_rbf_calibrator(dimension = 4)
mp_optimize(sir_cal)
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluation
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluation
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluation
#> $par
#> params params params params params params
#> 1.24623626 64.31809388 0.15367381 0.40557742 -0.11457659 0.33951668
#> params params
#> -0.07351877 0.61040344
#>
#> $objective
#> [1] 1601.412
#>
#> $convergence
#> [1] 1
#>
#> $iterations
#> [1] 126
#>
#> $evaluations
#> function gradient
#> 200 127
#>
#> $message
#> [1] "function evaluation limit reached without convergence (9)"
plot_fit(sir_cal)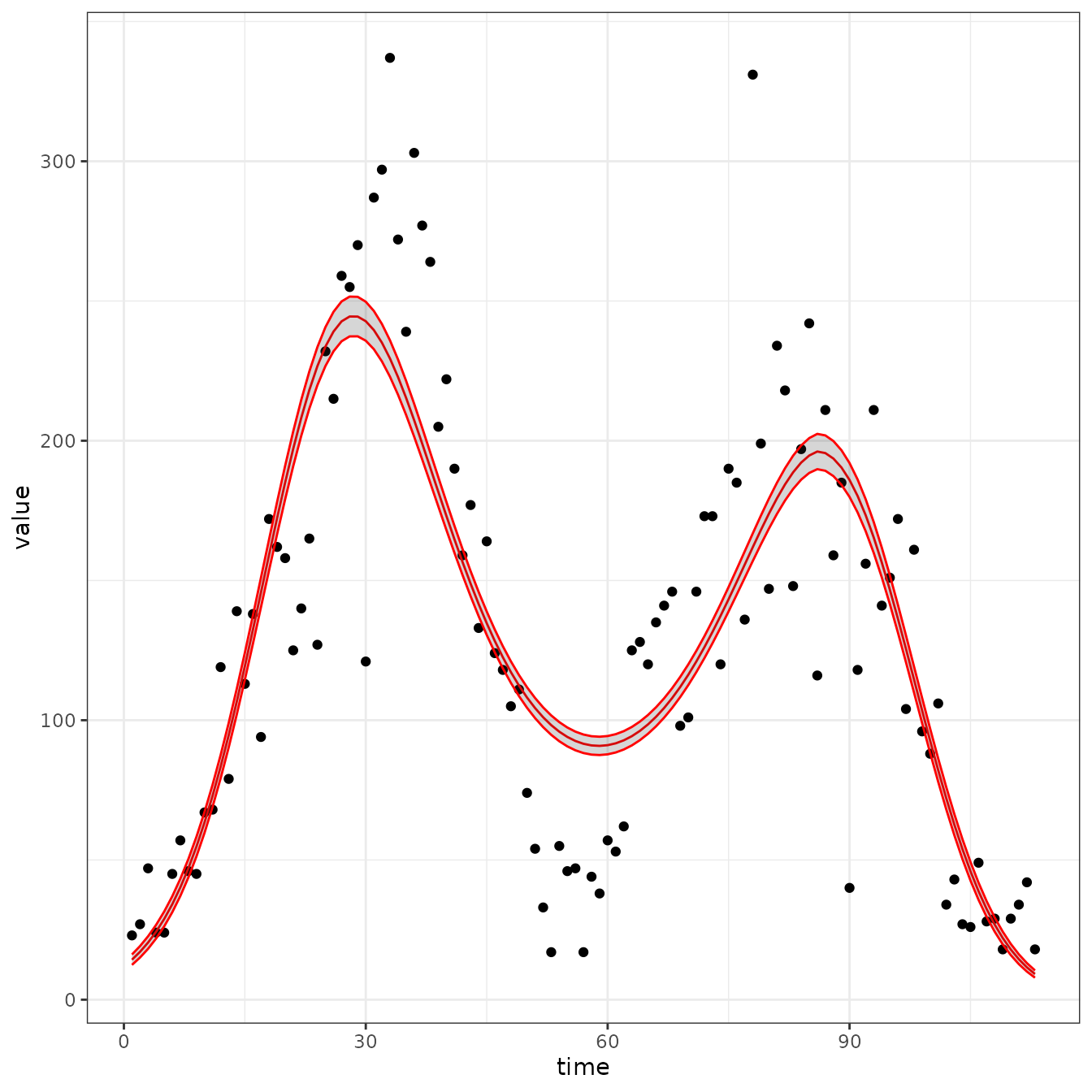
sir_cal = make_rbf_calibrator(dimension = 5)
mp_optimize(sir_cal)
#> $par
#> params params params params params params
#> -0.002793313 53.950823492 31.929825049 -4.573148593 1.944378326 -6.203399859
#> params params params
#> 1.785263470 -7.024005158 0.596297469
#>
#> $objective
#> [1] 1433.839
#>
#> $convergence
#> [1] 1
#>
#> $iterations
#> [1] 126
#>
#> $evaluations
#> function gradient
#> 200 127
#>
#> $message
#> [1] "function evaluation limit reached without convergence (9)"
plot_fit(sir_cal)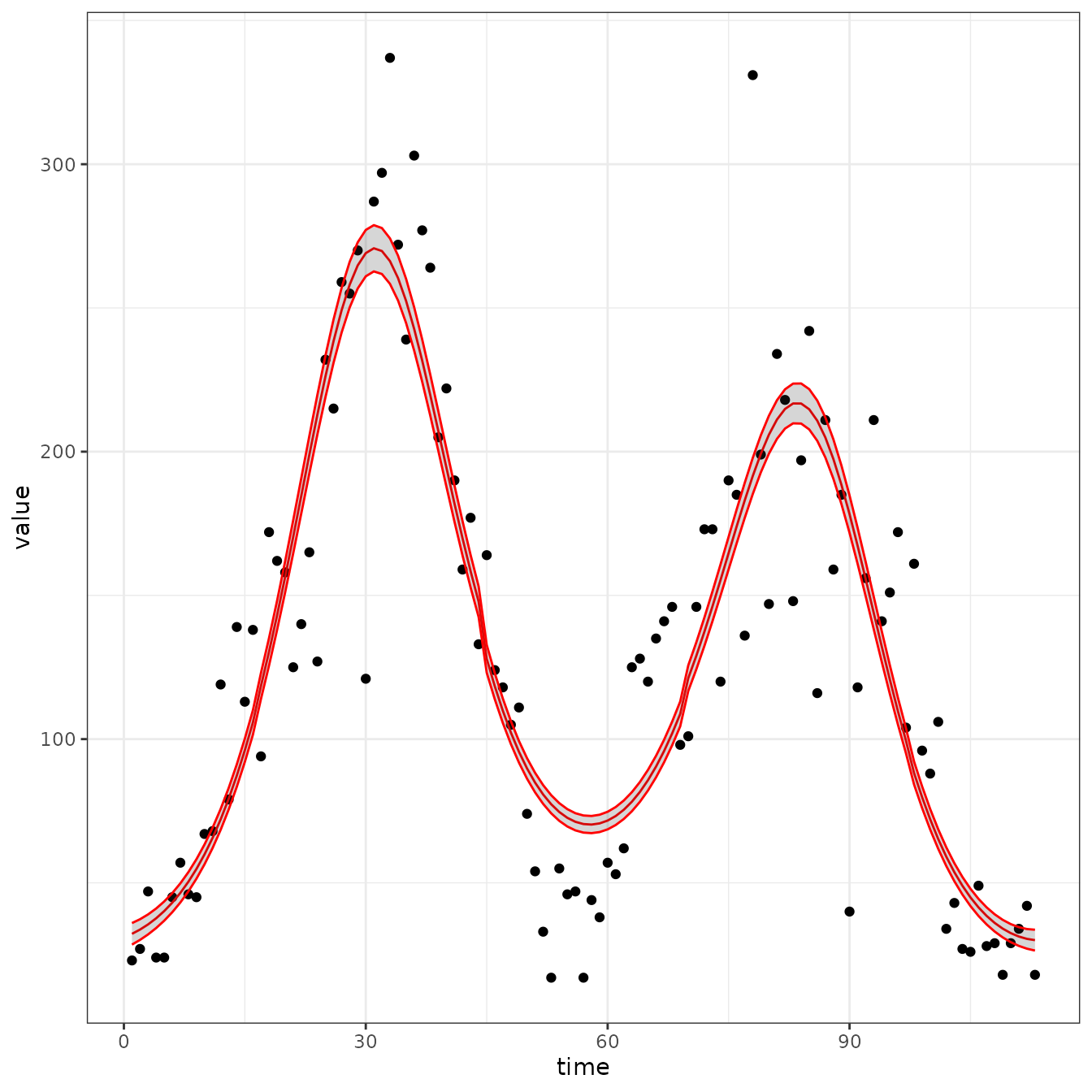
sir_cal = make_rbf_calibrator(dimension = 6)
mp_optimize(sir_cal)
#> Warning in (function (start, objective, gradient = NULL, hessian = NULL, :
#> NA/NaN function evaluation
#> $par
#> params params params params params params params
#> 0.7601931 8.1384172 5.0613008 -0.2998314 0.2581251 -0.6205325 0.3072368
#> params params params
#> -0.3905223 -0.1563821 0.5863554
#>
#> $objective
#> [1] 1441.671
#>
#> $convergence
#> [1] 1
#>
#> $iterations
#> [1] 120
#>
#> $evaluations
#> function gradient
#> 200 120
#>
#> $message
#> [1] "function evaluation limit reached without convergence (9)"
plot_fit(sir_cal)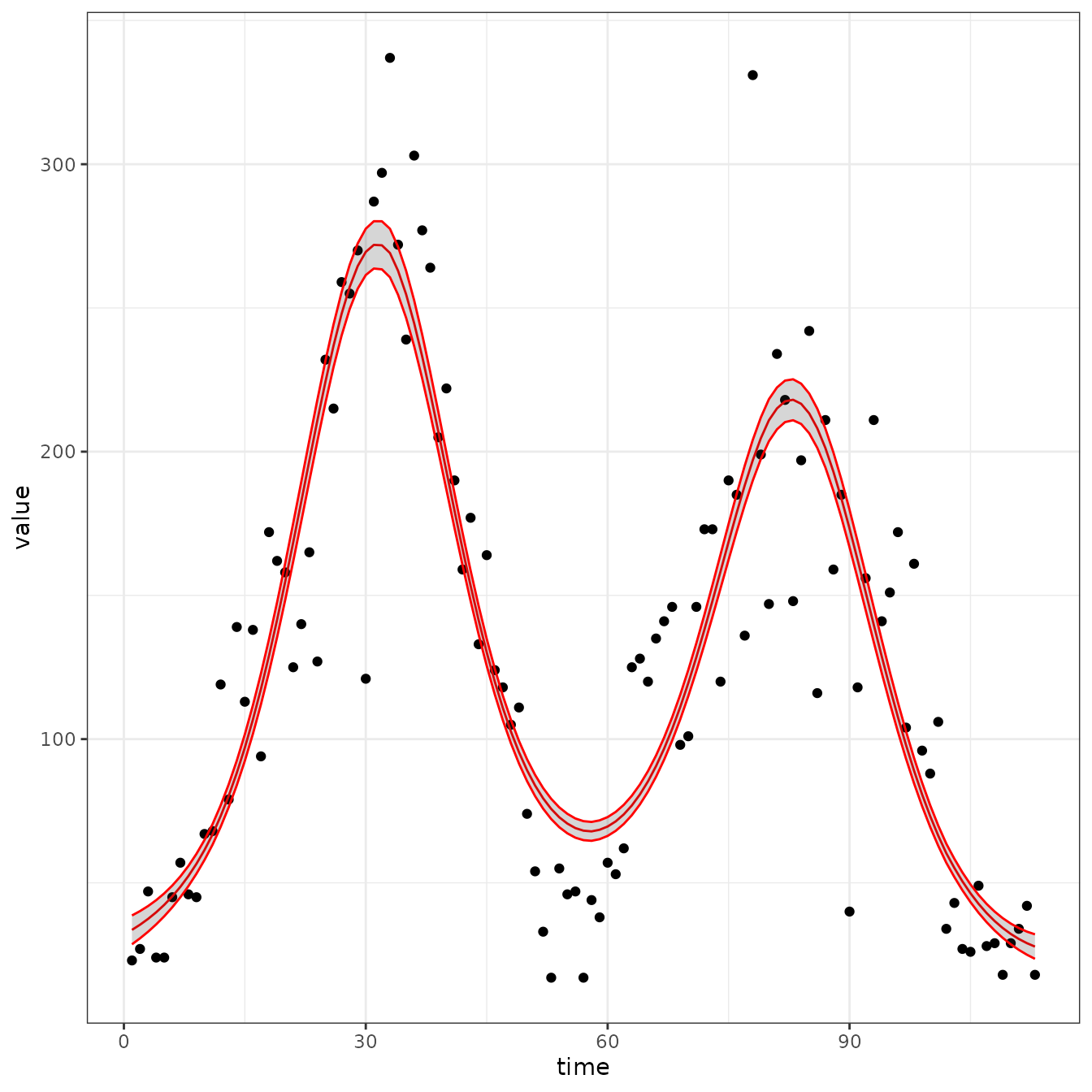
sir_cal = make_rbf_calibrator(dimension = 7)
mp_optimize(sir_cal)
#> $par
#> params params params params params params
#> 0.02862237 13.98682503 6.43589129 -0.64364609 -1.38502755 -0.17206581
#> params params params params params
#> -3.83455147 -0.06295877 -1.56407248 -3.70587627 0.58017538
#>
#> $objective
#> [1] 1471.266
#>
#> $convergence
#> [1] 1
#>
#> $iterations
#> [1] 117
#>
#> $evaluations
#> function gradient
#> 200 117
#>
#> $message
#> [1] "function evaluation limit reached without convergence (9)"
plot_fit(sir_cal)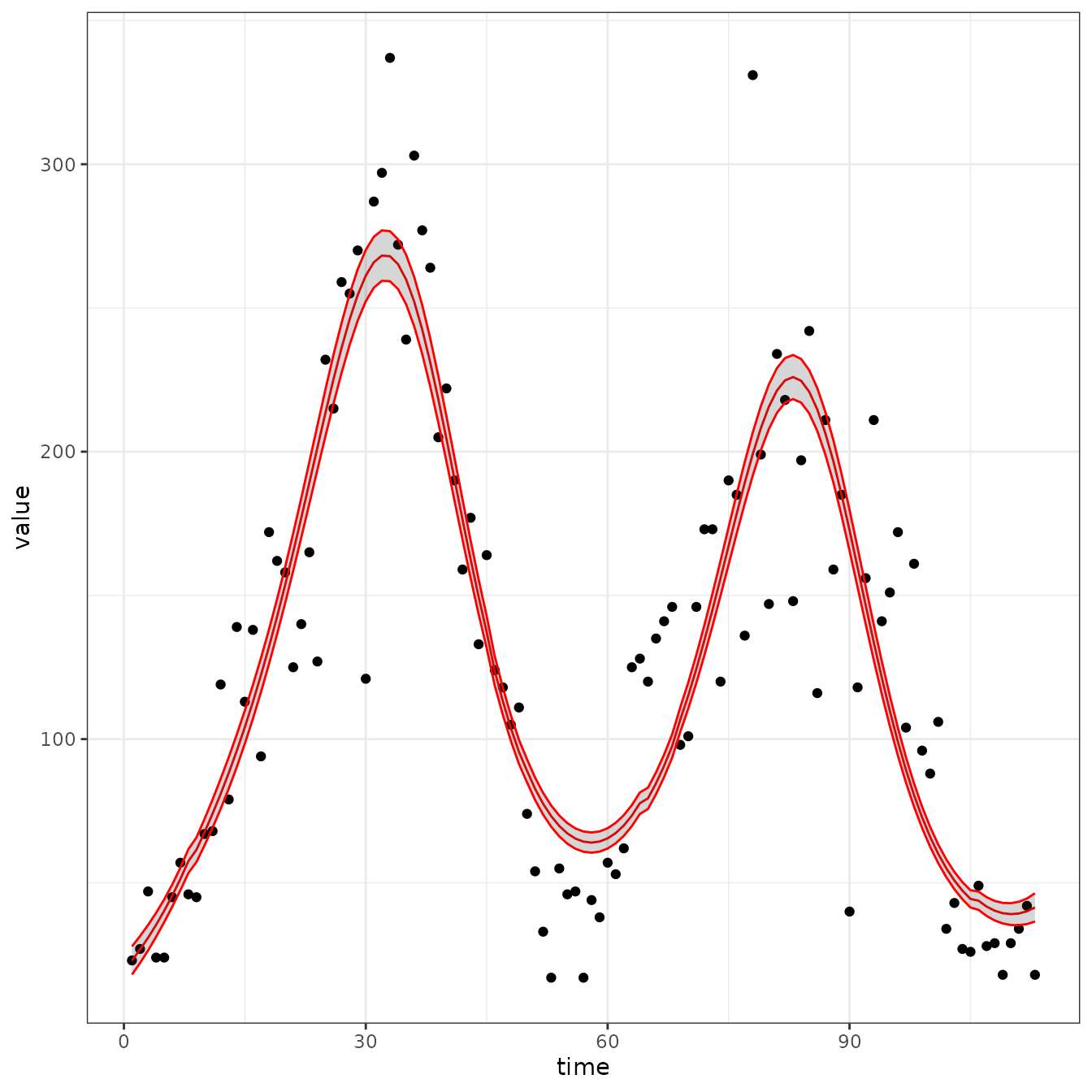
Interestingly we have now managed to fit gamma
mp_tmb_coef(sir_cal)
#> term mat row col default type estimate std.error
#> 1 params gamma 0 0 0.100000000 fixed 0.02862237 0.006524905
#> 2 params.1 I 0 0 1.000000000 fixed 13.98682503 18.333741358
#> 3 params.10 prior_sd_beta 0 0 1.000000000 fixed 0.58017538 0.018255583
#> 4 params.2 report_prob 0 0 0.003333333 fixed 6.43589129 8.439943861
#> 5 params.3 time_var_beta 0 0 0.000000000 fixed -0.64364609 0.183442769
#> 6 params.4 time_var_beta 1 0 0.000000000 fixed -1.38502755 0.149596026
#> 7 params.5 time_var_beta 2 0 0.000000000 fixed -0.17206581 0.118303816
#> 8 params.6 time_var_beta 3 0 0.000000000 fixed -3.83455147 0.138041586
#> 9 params.7 time_var_beta 4 0 0.000000000 fixed -0.06295877 0.166110832
#> 10 params.8 time_var_beta 5 0 0.000000000 fixed -1.56407248 0.105245714
#> 11 params.9 time_var_beta 6 0 0.000000000 fixed -3.70587627 0.244865141This is a much slower recovery rate. Expected time in the R box is about one year! Not believeable, but the point is to make it easier to fit models so you can try more things.
mp_tmb_calibrator(
spec = sf_sir
, data = observed_data
, traj = "reports"
, tv = mp_rbf("beta", dimension = 7)
, par = c("gamma", "I", "report_prob")
, outputs = c("reports", "beta")
)
#> ---------------------
#> Before the simulation loop (t = 0):
#> ---------------------
#> 1: S ~ N - I - R
#> 2: outputs_var_beta ~ group_sums(values_var_beta * time_var_beta[col_indexes_beta], row_indexes_beta, outputs_var_beta)
#> 3: outputs_var_beta ~ c(outputs_var_beta[0], outputs_var_beta)
#>
#> ---------------------
#> At every iteration of the simulation loop (t = 1 to T):
#> ---------------------
#> 1: beta ~ exp(time_var(outputs_var_beta, data_time_indexes_beta))
#> 2: infection ~ S * (beta * I/N)
#> 3: recovery ~ I * (gamma)
#> 4: S ~ S - infection
#> 5: I ~ I + infection - recovery
#> 6: R ~ R + recovery
#> 7: reports ~ infection * report_prob
#>
#> ---------------------
#> After the simulation loop (t = T + 1):
#> ---------------------
#> 1: sim_reports ~ rbind_time(reports, obs_times_reports)
#>
#> ---------------------
#> Objective function:
#> ---------------------
#> ~-sum(dpois(obs_reports, clamp(sim_reports))) - sum(dnorm(values_var_beta, 0, prior_sd_beta))
