Specifying Time-Varying Parameters
Source:vignettes/time-varying-parameters.Rmd
time-varying-parameters.RmdBaseline SIR Model
Here we modify an SIR model so that transmission rate is time-varying.
sir = Compartmental(system.file("starter_models", "sir", package = "macpan2"))
simulator = sir$simulators$tmb(time_steps = 50
, state = c(S = 99, I = 1, R = 0)
, flow = c(foi = 0, gamma = 0.2)
, N = empty_matrix
, beta = 0.8
)
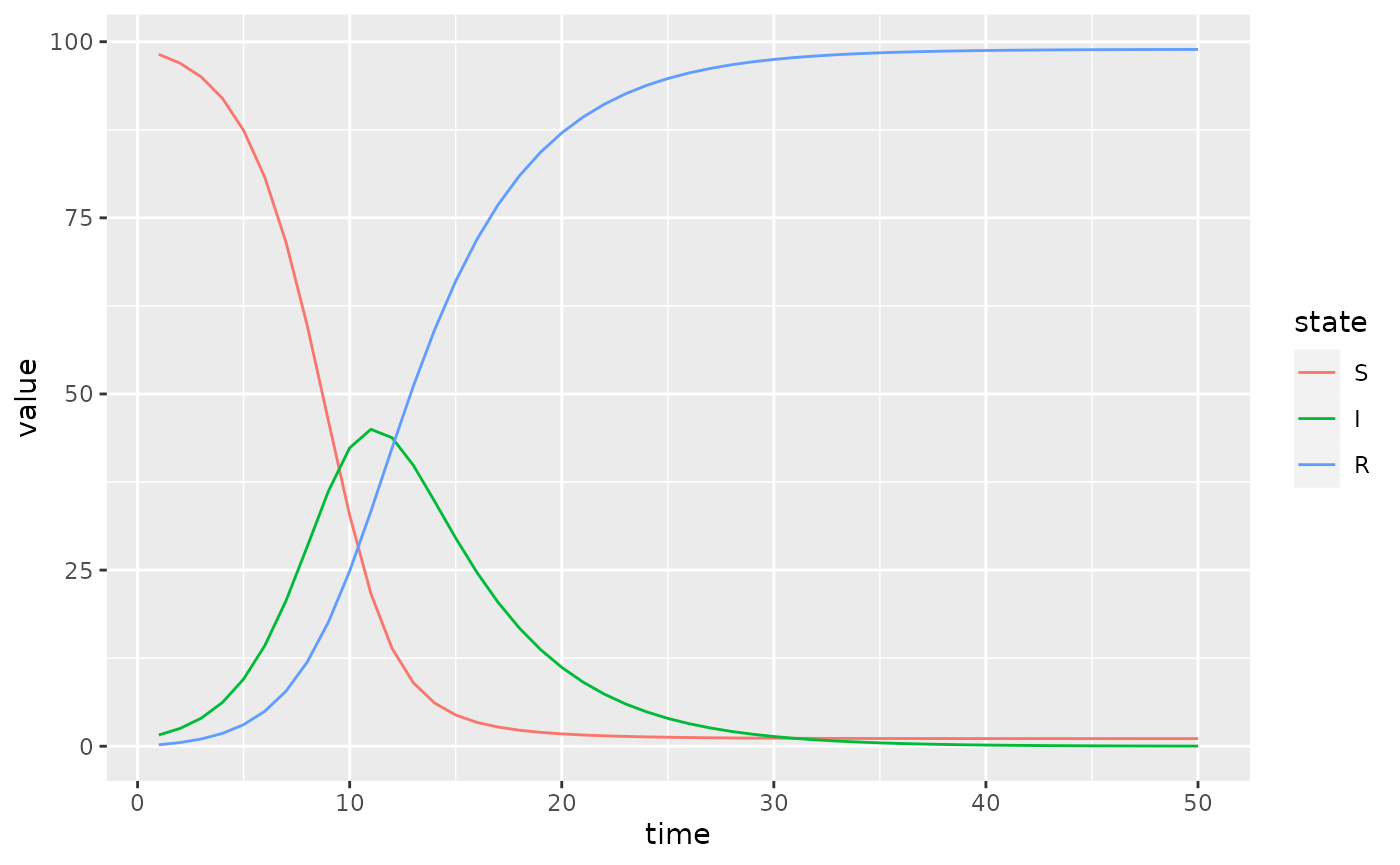
(simulator$report(.phases = "during")
%>% rename(state = row)
%>% mutate(state = factor(state, sir$labels$state()))
%>% ggplot() + geom_line(aes(time, value, colour = state))
)
Piecewise Time Variation
We now change the value of the transmission rate, beta,
at the beginning of time-step 10 and 15. In the first step we add to the
simulator a vector containing these change-points.
simulator$add$matrices(beta_changepoints = c(0, 10, 15))Next we add the values to which beta changes at these
time-steps.
simulator$add$matrices(beta_values = c(0.8, 0.01, 0.4))We also need a variable to track the current value of
beta. This beta_pointer starts at time-step
equal to 0, and it will be incremented throughout the simulation.
simulator$add$matrices(beta_pointer = 0)We increment beta_pointer using the time_group
function that returns either beta_pointer or
beta_pointer + 1 depending on whether or not the current
time-step is at a change-point in beta_changepoints.
simulator$insert$expressions(
beta_pointer ~ time_group(beta_pointer, beta_changepoints),
.phase = "during"
)We update beta at every iteration of the simulation loop
using this beta_pointer.
simulator$insert$expressions(
beta ~ beta_values[beta_pointer],
.phase = "during"
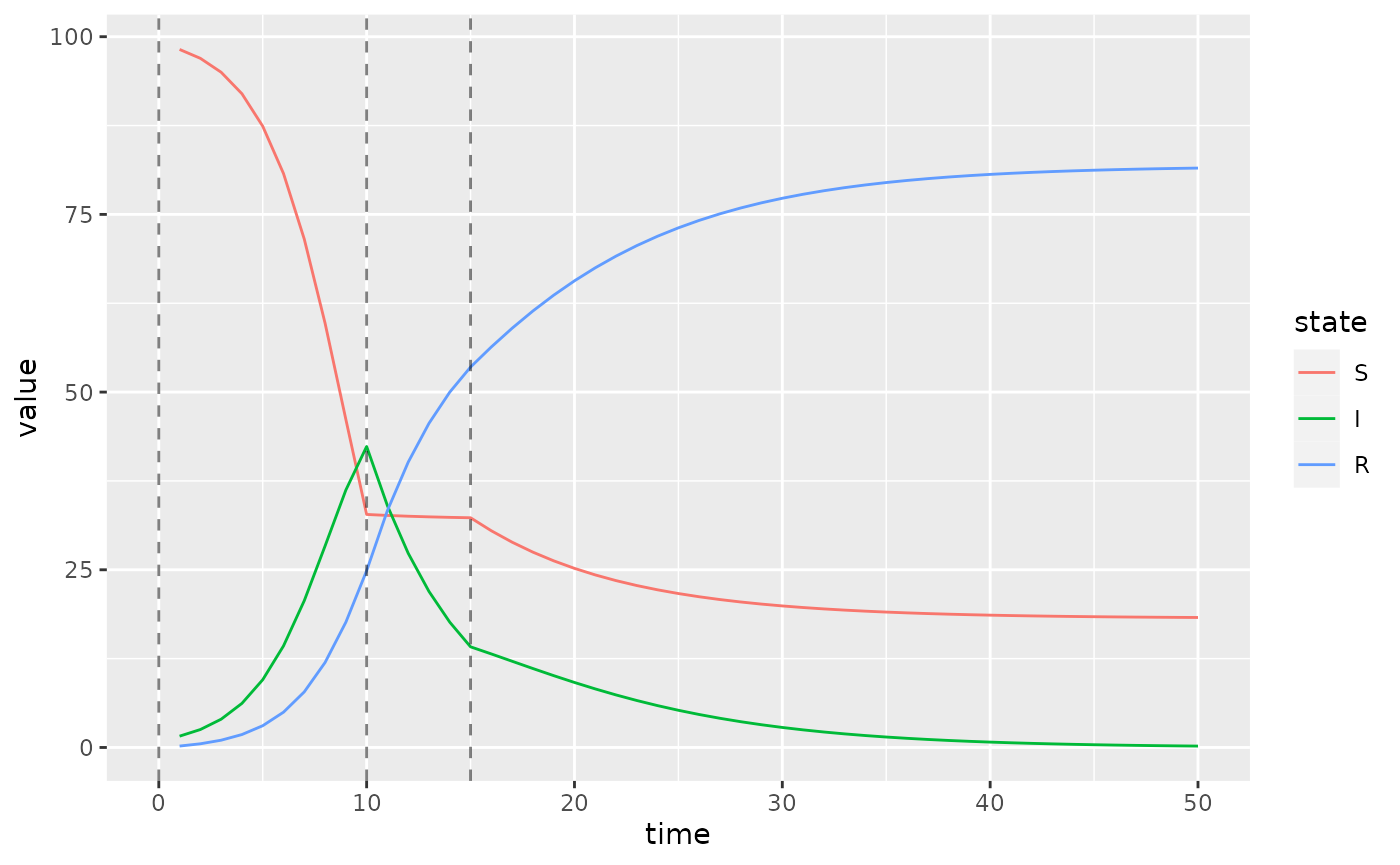
)And that’s it. Now we plot the updated simulations using these change-points, which we highlight with vertical lines.
s = simulator$report(.phases = "during")
(s
%>% rename(state = row)
%>% mutate(state = factor(state, sir$labels$state()))
%>% ggplot()
+ geom_line(aes(time, value, colour = state))
+ geom_vline(
aes(xintercept = x),
linetype = "dashed",
alpha = 0.5,
data = data.frame(x = simulator$get$initial("beta_changepoints"))
)
)
Calibrating Time Variation Parameters
First we simulate data to fit our model to, to see if we can recover the time-varying parameters.
set.seed(1L)
I_observed = rpois(
50,
filter(s, matrix == "state", row == "I")$value
)
plot(I_observed)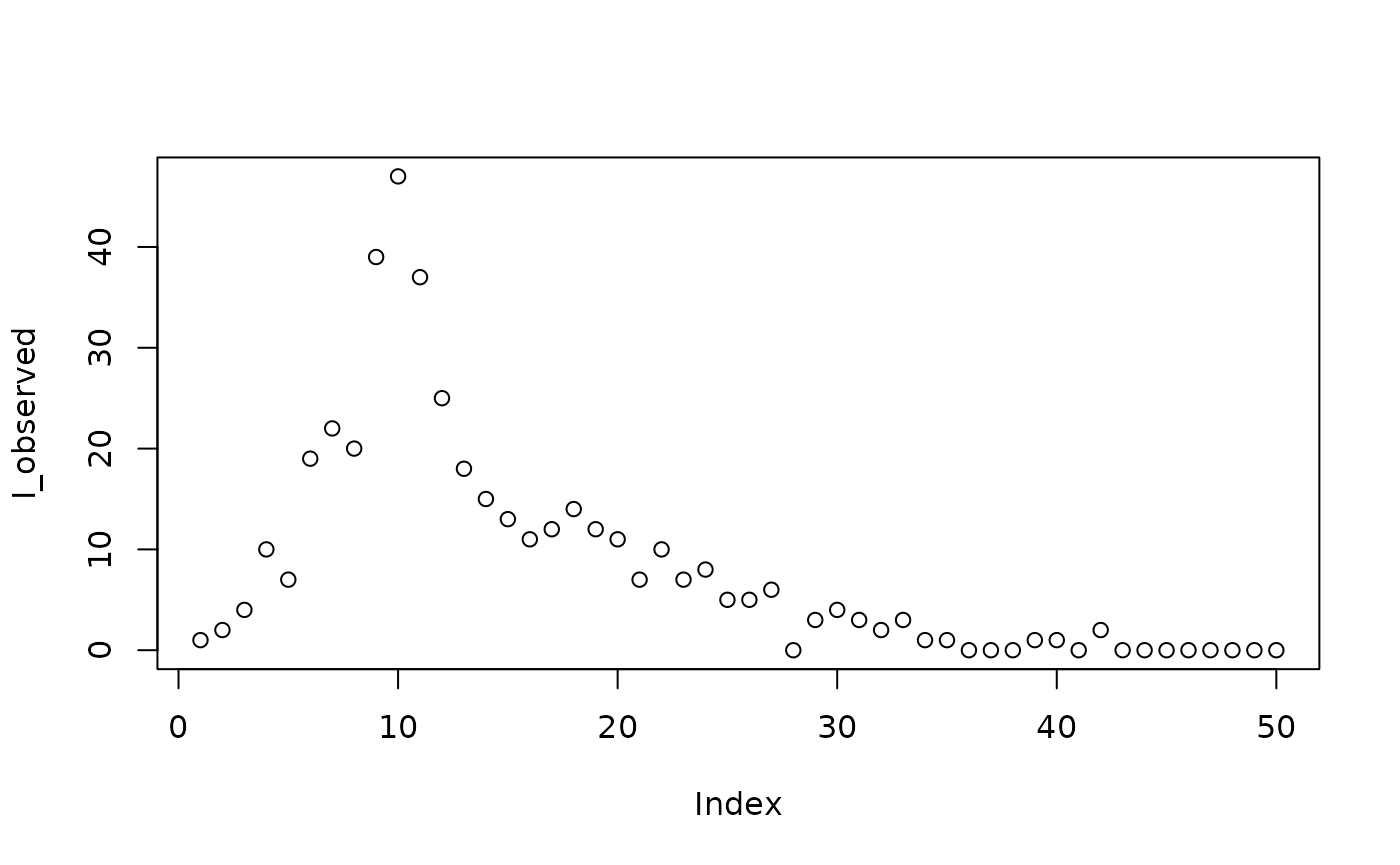
Then we add a few matrices to the model for keeping tracking of information used in model fitting.
simulator$add$matrices(
## observed data
I_obs = I_observed,
## simulated trajectory to compare with data
I_sim = empty_matrix,
## location of I in the state vector
## (the `-1L` bit is to get 0-based indices instead of 1-based)
I_index = match("I", sir$labels$state()) - 1L,
## matrix to contain the log likelihood values at
## each time step
log_lik = empty_matrix,
## need to save the simulation history of each of these matrices
.mats_to_save = c("I_sim", "log_lik")
)Now we need some new expressions. The first expression pulls out the I state from the state vector.
simulator$insert$expressions(
I_sim ~ state[I_index],
.phase = "during"
)The second expression computes a vector of Poisson log-likelihood values – one for each time step.
simulator$insert$expressions(
log_lik ~ dpois(I_obs, clamp(rbind_time(I_sim))),
.phase = "after"
)
simulator$replace$obj_fn(~ -sum(log_lik))Next we declare the beta values as parameters to be optimized on the log scale.
simulator$add$transformations(Log("beta_values"))
simulator$replace$params(
default = log(mean(simulator$get$initial("beta_values"))),
mat = rep("log_beta_values", 3L),
row = 0:2
)Finally we fit the model back to the simulation data.
simulator$optimize$nlminb()
#> Constructing atomic D_lgamma
#> outer mgc: 736.1414
#> Constructing atomic D_lgamma
#> outer mgc: 112.6655
#> outer mgc: 71.38648
#> outer mgc: 13.6616
#> outer mgc: 12.58091
#> outer mgc: 3.638083
#> outer mgc: 1.399513
#> outer mgc: 1.609453
#> outer mgc: 1.140347
#> outer mgc: 0.5870909
#> outer mgc: 0.2527136
#> outer mgc: 0.09938628
#> outer mgc: 0.03753935
#> outer mgc: 0.01394885
#> outer mgc: 0.005150654
#> outer mgc: 0.001897432
#> outer mgc: 0.0006983805
#> outer mgc: 0.0002569678
#> outer mgc: 9.453969e-05
#> outer mgc: 3.478009e-05
#> outer mgc: 1.2795e-05
#> outer mgc: 4.707033e-06
#> outer mgc: 1.731623e-06
#> outer mgc: 6.370286e-07
#> outer mgc: 2.3435e-07
#> outer mgc: 8.621247e-08
#> outer mgc: 3.171579e-08
#> $par
#> params params params
#> -0.1710158 -22.8268369 -0.8291142
#>
#> $objective
#> [1] 99.74231
#>
#> $convergence
#> [1] 0
#>
#> $iterations
#> [1] 26
#>
#> $evaluations
#> function gradient
#> 28 27
#>
#> $message
#> [1] "relative convergence (4)"We can see that the optimizer converges
(i.e. $convergence = 0) in 26 iterations.
On the log scale we see that the optimizer finds different values
(current) than it started at (default).
simulator$current$params_frame()
#> par_id mat row col default current
#> 1 0 log_beta_values 0 0 -0.9079919 -0.1710158
#> 2 1 log_beta_values 1 0 -0.9079919 -22.8268369
#> 3 2 log_beta_values 2 0 -0.9079919 -0.8291142More importantly the beta values on the untransformed scale recover to the values used in the simulations, although the second fitted beta value is much smaller than the true value.
data.frame(
fitted = formatC(
exp(simulator$current$params_frame()$current),
format = "e", digits = 2
),
true = simulator$get$initial("beta_values")
)
#> fitted true
#> 1 8.43e-01 0.80
#> 2 1.22e-10 0.01
#> 3 4.36e-01 0.40